 cancer-sim
cancer-sim 
 cancer-sim
cancer-sim 
Tissue displacement from treatment of high-grade glioma as described by non-rigid registration or a fitted radial deformation model of tissue expansion or shrinkage.
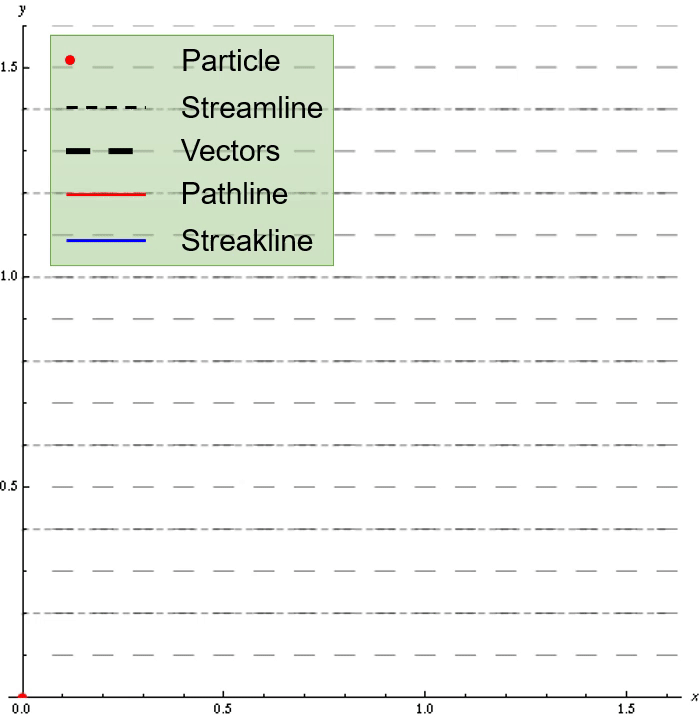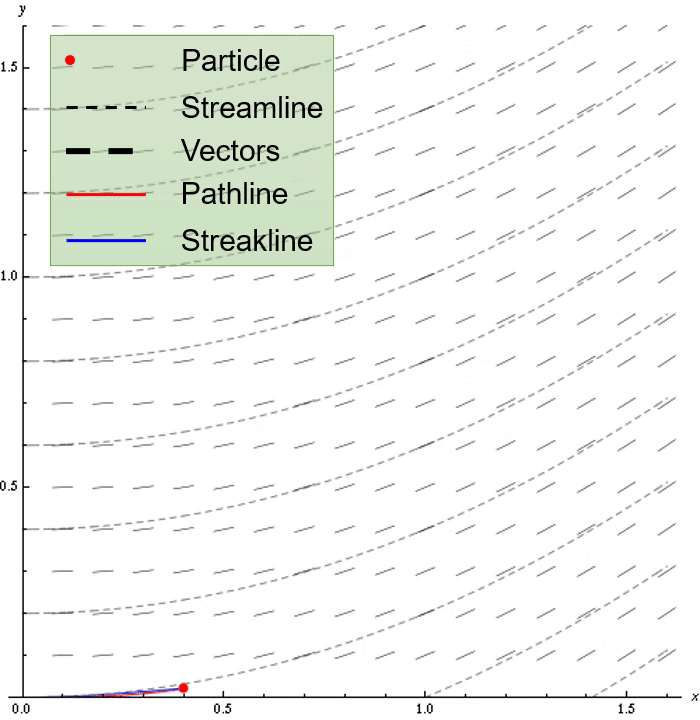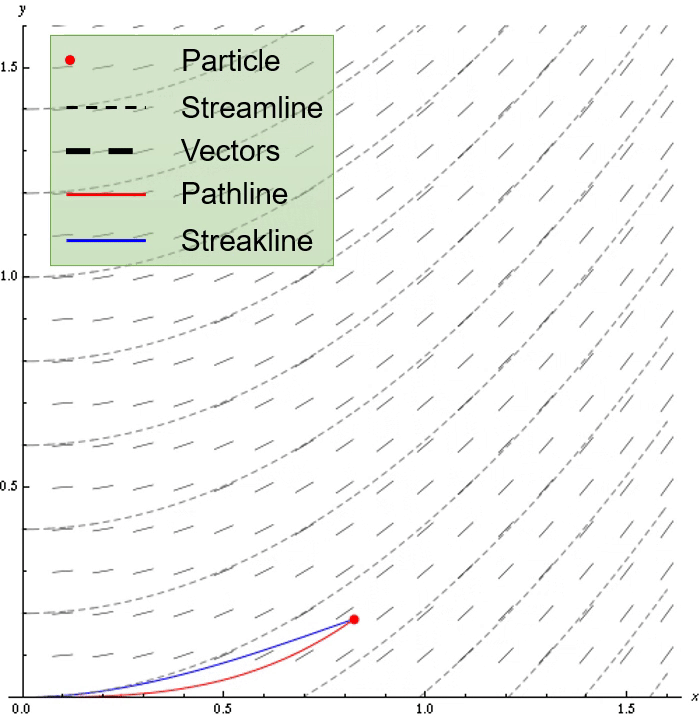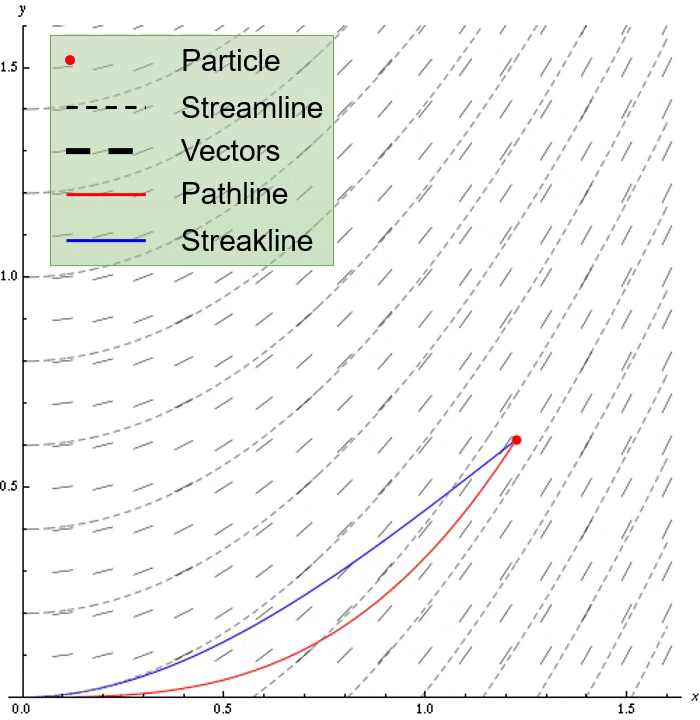

Non-rigid registration methods can be used to estimate tissue displacement between MRI examinations. Our results indicate applicability of non-rigid registration for tumor growth characterization for low physical displacements (~3 mm), large tumors (high infiltration) and contrast-enhanced and edematous tissue. The highest performant methods were ANTs Symmetric image Normalization (SyN) with cross-correlation as loss metric and Gunnar-Farneback optical flow.
The radial deformation model was developed to validate the applicability of non-rigid registration for tumor growth characterization. It takes as input a structural MRI scan, lesion and brain masks, and three parameters:
Expanding deformations mimic tumor progression with a pushing phenotype from cancer mass effect, while shrinking deformations mimic shrinkage of pathogenic tissue as a result of treatment. Grid search on the parameters were used to find best fit model projections for longitudinal MRI data.
The core idea of the deformation model is illustrated in the axial brain slice figure below for expanding deformation. Based on the bounding box dimensions of a lesion mask, an outward vector field from a 3D gaussian intensity function is computed. To model infiltration, the field is "stretched out" by interpolating the field components in the double dashed region region to cover the entire single dashed region. The field is then scaled to have the specified maximum tissue displacement. An optional Perlin noise can be added to mimic displacement irregularities.

For review (2021-05-03)
Ivar Thokle Hovden
PhD Student
Department of Diagnostic Physics
Computational Radiology and Artificial Intelligence (CRAI)
Division of Radiology and Nuclear Medicine
Oslo University Hospital (OUS)
Department of Physics
University of Oslo (UiO)
Norway
Home page at OUS
Home page at UiO
GitHub

Mattis Hovden Aas, Dovre Media